| –ù–∞–∑–≤–∞–Ω–∏–µ | : | Setup RNA-Seq Pipeline from scratch: fastq (reads) to counts | Step-by-Step Tutorial |
| –ü—Ä–æ–¥–æ–ª–∂–∏—Ç–µ–ª—å–Ω–æ—Å—Ç—å | : | 31.50 |
| –î–∞—Ç–∞ –ø—É–±–ª–∏–∫–∞—Ü–∏–∏ | : | |
| –ü—Ä–æ—Å–º–æ—Ç—Ä–æ–≤ | : | 68¬Ýrb |




|
|
Hello there It is a very informative video kindly make a tutorial video of miRNA processing from fastq to readcount Thanks Comment from : @mahmad6210 |
|
|
Thank you so much for this video it's very helpfulbrCan you please make a video of further steps like count matrix and finding deg's Comment from : @nk12987 |
|
|
Hi! Wonderful videomakes things so easy to understand I had one question though After alignment when do we use Cufflinks and when to use featureCounts? What differences are there in the two tools? First one being cufflinks-cuffmerge-cuffdiff and the latter being featureCounts-Deseq/EdgeR Comment from : @debashruti4984 |
|
|
I don't have the genome sequencing dataI need to do de novo assembly of my transcriptome datacan you please elaborate how can I do this? What additional steps I need to follow before hisat2? Comment from : @sutrishakundu2147 |
|
|
Thank you!!! You've been a lifesaver for me as a newbie to the lab Would love to see the ATAC-seq version of this ! Comment from : @wolveshowlhigh |
|
|
if i have replicates, at which step i will have to merge the two rna seq data? Comment from : @mohdfahad6224 |
|
|
Very helpful videoThank you! Comment from : @juniorkika254 |
|
|
Teach us with R Comment from : @aliaslam3303 |
|
|
Am unable to understand Comment from : @MD_waseem1 |
|
|
Maam why raw sequence number and after adapter removal sequence number varies as all raw read have adapter Comment from : @sonalidey1454 |
|
|
The link to the raw reads is not attached in the description Kindly provide please Comment from : @quinattasneemrafique536 |
|
|
Can you please make a detailed video on Metagenomics Comment from : @quinattasneemrafique536 |
|
|
Did you listen your presentation? Comment from : @andrewzlobin7011 |
|
|
Hey, I wish to quantify human micro RNAs from double stranded fastq files Which reference shall I use? Comment from : @shreyaslabhsetwar9055 |
|
|
Hi, quick question, the count table display gene ID's and their counts, how do I get more gene information? (such as gene name, symbol, and annotation added ) Thanks Comment from : @seiftekaya3212 |
|
|
@Bioinformagician can you do an Orthofinder tutorial ? Comment from : @linkandash8025 |
|
|
Great explanation for a beginner to get started with RNA seq! Cheers to you! Comment from : @sivanim3830 |
|
|
Great and clear… Thanks I got two questions though: 1 what’s the difference between the count matrix and TPM/FPKB/RPKB? Noting that TPM/FPKB/RPKB can’t be use for DEG, while count matrix can 2 Why Does bowtie can’t be used for alignment to genome? Comment from : @user-ns7ng2hz9w |
|
|
Great and clear… Thanks I got two questions though: 1 what’s the difference between the count matrix and TPM/FPKB/RPKB? Noting that TPM/FPKB/RPKB can’t be use for DEG, while count matrix can 2 Why Does bowtie can’t be used for alignment to genome? Comment from : @user-ns7ng2hz9w |
|
|
Thanks for the nice and informative session Comment from : @kiplimosimon1429 |
|
|
Thank you so much! Comment from : @sdmasroor |
|
|
Thanks for your beautiful videos! Can you make a similar video for PE reads and include the step of generating genome index files using HISAT2? That would be so helpful Comment from : @nayanbhowmik5010 |
|
|
how to download the tools required for the pipeline on a M2 MacBook? Comment from : @JaskaranSingh-om5mv |
|
|
can we convert fasta instead of fastq to count data? Comment from : @zulfakhan2655 |
|
|
How to use featureCounts command for paired end reads? and hisat2 as well? Comment from : @deepanshugarg4257 |
|
|
I just want to say thank you very much for doing this, im just starting out in my PhD studies and you have been really helpful! Comment from : @josefinagarciadiaz478 |
|
|
‚ù§ Comment from : @kruthirao1 |
|
|
Hi madam, how to convert raw datagz file to fpkm file You have used in your other video fpkn file and proceseed later using deseq2 library There you have taken directly tab file Buti have fastpnotab file How i should convert yhis to fpkm tab file Comment from : @grsbiosciences |
|
|
Brilliantly done! Comment from : @histephenson007 |
|
|
You're awsomee❤❤ Comment from : @tarahsify |
|
|
This is so clear--love your content Comment from : @apedike |
|
|
Hi, thanks for the video! But not all the commands are working I am using the terminal in macOS and changed to the bash shell What's the problem and how to fix it? Comment from : @user-mn4gu7kv2d |
|
|
Hi can someone guide me about the error: Failed to open the annotation file /celegan/caenorhabditisgtf, or it’s format is incorrect or it contains no ‘exon’ features Comment from : @Tekofilic |
|
|
please make a video on De Novo RNA seq with paired end reads - its a hot topic Comment from : @adeelabbasi7375 |
|
|
Please make a video of setting up a pipeline using Trinity for denovo sequence assembly Comment from : @subarnathakur6973 |
|
|
Hi, wonderful video so far (I'm only a little over halfway through) I was wondering why you chose to trim it by 10 base pairs? Was that just an arbitrary decision or is it recommended? Thank you!! Comment from : @HopefulHopelessly |
|
|
I run another dataset of rnaseq and the results of fastqc for this dataset is the same but with Sequence Duplication levels with a flag, this is significant in experiment of Rnaseq that analyze control vs treatment? Comment from : @gcbicca |
|
|
That was awesome ! Thanks so much Comment from : @gustavotamasco4914 |
|
|
When will you release the video about post alignment qc? :) Comment from : @tolga1292 |
|
|
Hi, great tutorial, i have a question I have aligned with star, bulk RNAseq from voxels clones, and i have 52 uniquely mapped reads, 30 of multimapped, and the rest of not aligned Can you explain different aspects of the output of star, if they have good quality or not? with FASTQC i cant find any adapter Comment from : @maytelopez-cascales6113 |
|
|
can you make a tutorial for NEXTFLOW or snakemake? Comment from : @mostafaismail4253 |
|
|
Hi I have watched couple of your videos and they are so informative! For this pipeline, can I ask if there are R packages that can do the same thing? bc I am not familiar with linux or bash scripts but I will be learning them Thanks for all the efforts you put in the videos Comment from : @ynpan910 |
|
|
You are doing such a great job ! Kudos! Would be willing to show the analysis of 10x spatial transcriptomics in the future ? Comment from : @rashmitan6867 |
|
|
So after this, how to analyze it on R? Comment from : @juliangrandvallet5359 |
|
|
Hi, this video is very concise Thanks for sharing I'm still struggling install HISAT2 onto M1 mac How did you do it? Comment from : @user-uq3qh2cy9v |
|
|
How can you make RNA-Seq analysis look so simple! :) Kudos great work! Comment from : @AjayJoshi-yl5zx |
|
|
Thank you for your video it was very helpful to learn about rna-seq workflow just quick questions: after hisat2 (alignment) is it essential to do variant calling stage before read counts? or I can just do readcounts using featurecount or htseq-count without process of SNP Comment from : @freezingtolerance7493 |
|
|
thank you for your video brI have a question : Can we use exactly the same pipeline to analyse single cell RNA sequencing data or it's not the same ?? brthank u in advance Comment from : @nourarifi2642 |
|
|
Wow, you really illustrate the workflow so systematically As a beginner, I only knew a little about RNA-seq analysis, so this helps a lot! Besides, you mention and explain lots of practical details, unlike some other video providers I gain so much from your videos! Many thanks to you!!! Comment from : @CHENYanZhen |
|
|
Load annotation file Homo_sapiensGRCh38108gtf ||
br|| Features : 1642501 ||
br|| Meta-features : 62703 ||
br|| Chromosomes/contigs : 47 ||
br|| ||
br|| Process BAM file SRR5205331bam ||
brERROR: Paired-end reads were detected in single-end read library : /mnt/c/Desktop/Bioinformatic/BC_Project/SRR5205331bambrbrI am getting this error WHy? Comment from : @motiarrahman9160 |
|
|
Brilliant! Thank you very much for the tutorial! Comment from : @felipevilicich980 |
|
|
Are you using R for this tutorial ? Comment from : @stanyang4321 |
|
|
Thank you very much for this wonderful video Keep it up I was also able to perform most of these tasks in Galaxy Comment from : @kingsleyibeabuchi3461 |
|
|
Great job Your vidoes have been very helpful I am sincerely grateful Comment from : @kingsleyibeabuchi3461 |
|
|
Hi! Great video I was wondering if there is a similar quality check for CEL files Do you happen to know? Comment from : @recordinghoney |
|
|
I really like your videos This tutorial help me go over the bumpy learning curve! I am so excited that I can get the read count by myself Thanks for putting your time to make these videos Comment from : @chienlee4695 |
|
|
thank you, its really informative for me Comment from : @ashishbhaladhare |
|
|
Thanks a lot for this tutorial, it helps me a lot to start with What's your math background? Have you studied advanced statistic or something related to it? Comment from : @francescosilvestro2092 |
|
|
Thank you so much for this helpful videosbrplease keep making such a great contentbrand I have one question, when are you going to upload a video covering FastQC analysis Comment from : @yaarobaltali6076 |
|
|
It was really helpful ma'am But you didn't explain how to download featurecounts None are being able to explain it properly I am stuck now ü•≤ Comment from : @pradyutdas7358 |
|
|
Thank You ! Comment from : @kajalpanchal8239 |
|
|
hello, thank you for your videos, and can you please make video about the github, so uploading and presenting the projects on github? Comment from : @serrasonmez7912 |
|
|
Good show, thx a lot Comment from : @renqiuguoli1258 |
|
|
Thank you so much Very understandable and easy to follow Comment from : @christianahoyindamolajemiy5531 |
|
|
It was a wonderful video Comment from : @ritikabassi1998 |
|
|
Thanks for this tutorial!brbrCould u help me on how can i create an index based on other genome like Aedes aegypti
bri want to make RNA-seq analysis of infected mosquitoes, wich file should i use as reference genome ? Comment from : @guilhermecardosodevargas4762 |
|
|
You are such a blessing Comment from : @Gbemi78 |
|
|
Thank you very much for this video!brI have a question for paired end data After doing samtools sort, the reverse read and the forward read are not consequetive reads in the sorted bam anymore Is that okey? Comment from : @sofiapuvogel3663 |
|
|
Could you please make a video of how to trans counts data into FPKM or RPKM or TPM? Comment from : @anxu4239 |
|
|
This is really helpful Comment from : @arpanparichha6784 |
|
|
You're very sweet Comment from : @terryadams2652 |
|
|
Thank you, thank you for these wonderful videos! Perfect for beginners and they've helped immensely for completing my thesis! Comment from : @Luvinlife411 |
|
|
Great video I was wondering when are you planning to show the next steps From count matrix to DEA Comment from : @acramulhaquekabir5852 |
|
|
thank you, girl Comment from : @rafaelamachadotugores4769 |
|
|
Thanks for your video It is amazing Comment from : @ishakyusuf4616 |
|
|
Hi Bioinformatician I use STAR instead of hisat2 so what command I should use to run your data? Comment from : @chrisdoan3210 |
|
|
Hi Bioinformatician Would you please tell me why we have many pipelines for RNA-Seq? So these steps below are not considered pipeline (workflow):br1 Check quality with FastQCbr2 Trim reads with Trimmomaticbr3 Align reads to the reference with STARbr4 Calculate gene hit counts with FeatureCountsbr5 Compare hit counts between groups with DESeq2 Comment from : @chrisdoan3210 |
|
|
Great informational video Thanks a tonüôè If possible please make a video on de novo Trinity pipeline Comment from : @rahultanpure1338 |
|
|
Would you please guide me on how to set up the tools in this tutorial for a Mac M1 user because I got a lot of difficulties and errors for about a month? Thank you so much! Comment from : @chrisdoan3210 |
|
|
Thank you for this video !! Comment from : @tushardhyani3931 |
|
|
Thank you for this tutorial! I hope that you will create a patreon to support your channel :) Comment from : @CostanzoPadovano |
|
|
You r doing better than other Keep it up Comment from : @sandeepbansalCDRI |
|
|
Do you plan to have a similar pipeline in R ? Comment from : @mohammedfarhanlakdawala4326 |
|
|
Amazing easy to follow video! I was Just wondering if you could clarify how the lines of code would change for paired ends reads, specifically when using the TruSeq Stranded mRNA kit for bulk RNA-seq library prep Thanks!! Comment from : @louisepitcher6180 |
|
|
Thank you for your videos! When I run this code: hisat2 -q --rna-strandness R -x HISAT2/grch38/genome -U data/demo_trimmedfastq I got error message: hisat2: command not found Would you tell me how to fix this? Does the way we install hitsat2 matter? What is location of hisat2 should be? Comment from : @chrisdoan3210 |
|
|
Can please post a tutorial for vcf file analysis with R Comment from : @ShivangiSinghMBB |
|
|
Youtube is a library You are one of the books that I frequently read Thanks! :-) Comment from : @kitdordkhar4964 |
|
|
Good job!!! Comment from : @rushupatel |
|
|
Amazing helpful video Congratulations!!! Comment from : @alvaroruiztabas5627 |
|
|
cannot be happier <3 Comment from : @escastorage7427 |
|
|
Thanks for this great tutorial! brQuestion - If you have high duplicates rate (say 70-80 in case of FFPE) pre alignment, would you remove them? brbrThere are some wet lab QC metrics like DV200, RIN, etc that also determines the effectiveness of downstream analysis might be worth considering/including for your tutorial on QCbrKeep it up and Cheers! Comment from : @amitrupani9898 |
|
|
Great mam I need help for building cnv pipeline will be a great help if u tell anyway for contacting you Comment from : @kathirvelm755 |
|
|
Awesome, thank you for doing this video [much awaited] -- so nice of you Khushbu -- Please keep making such great stuff -- Many thanks Comment from : @aamirmalik7740 |
|
|
You’re the best ! Comment from : @shrishteekandoi120 |
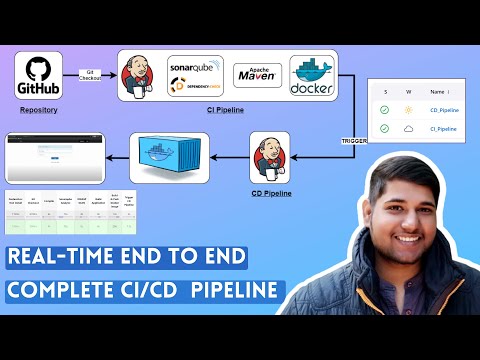 |
Real-Time CI CD Pipeline Project | CI CD Pipeline | Jenkins CI CD Pipeline –Ý—ï–°‚Äö : DevOps Shack Download Full Episodes | The Most Watched videos of all time |
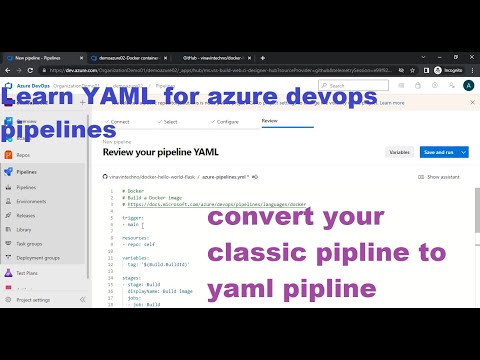 |
Azure Devops YAML CI Pipeline | Learn YAML for Azure devops pipeline | classic pipeline to YAML –Ý—ï–°‚Äö : Avin Techno Download Full Episodes | The Most Watched videos of all time |
 |
Build CI and CD Pipeline using Azure DevOps - Step by Step | AZ-400:Azure DevOps | CICD Pipeline –Ý—ï–°‚Äö : BestDotNetTraining Download Full Episodes | The Most Watched videos of all time |
 |
Jenkins CICD Declarative Pipeline English | How To Create Declarative Pipeline Step By Step English –Ý—ï–°‚Äö : ExploreDevops Download Full Episodes | The Most Watched videos of all time |
 |
SPSS Tutorial 8 - Combining Percentages and Frequency Counts in Cross-tabulations –Ý—ï–°‚Äö : patrickkwhite Download Full Episodes | The Most Watched videos of all time |
 |
4. Jenkins Pipeline - Build and Deploy a war file on Tomcat Server | Pipeline script for CI/CD –Ý—ï–°‚Äö : Valaxy Technologies Download Full Episodes | The Most Watched videos of all time |
 |
37. Run Synapse notebook from pipeline | Pass values to Notebook parameters from pipeline in Synapse –Ý—ï–°‚Äö : WafaStudies Download Full Episodes | The Most Watched videos of all time |
 |
How to run selenium tests in Azure DevOps pipeline | Create Pipeline to run automation test cases | –Ý—ï–°‚Äö : Viren Automation Testing Download Full Episodes | The Most Watched videos of all time |
 |
Efficient Pipeline Repairs with Pipeline Leak Repair Clamps –Ý—ï–°‚Äö : DWT Pipe Tools Download Full Episodes | The Most Watched videos of all time |
 |
Betaflight Setup u0026 Tutorial - Step by Step Guide ? + Gps Setup! –Ý—ï–°‚Äö : Drone Camps RC Download Full Episodes | The Most Watched videos of all time |